Global Vaccinations
Faceted geospatial map of vaccines used by countries.
Today we will make the faceted geospatial charts from the article Tracking Coronavirus Vaccinations Around the World and find out about a bug relating to faceting geospatial charts and how to go about achieving the plot till a proper fix is made.
#hide_output
import pandas as pd
import altair as alt
alt.renderers.set_embed_options(actions=False)
owd_vaccine_uri = "https://github.com/owid/covid-19-data/blob/master/public/data/vaccinations/locations.csv"
vaccine_uri_raw_data = "https://raw.githubusercontent.com/owid/covid-19-data/master/public/data/vaccinations/locations.csv"
raw_data = pd.read_csv(vaccine_uri_raw_data)
raw_data.tail()
Let's study what unique vaccines are there -
from itertools import chain
set(chain(*raw_data.vaccines.apply(lambda x: map(str.lstrip, x.split(','))).reset_index(drop=True)))
Now we will split vaccines into a list and explode it so that the dataframe is in the correct format for our visualization purposes -
vaccine_location_data = raw_data.copy()
vaccine_location_data['vaccines'] = vaccine_location_data.vaccines.apply(lambda x: list(map(str.lstrip, x.split(','))))
vaccine_location_data = vaccine_location_data.explode('vaccines').reset_index(drop=True)
vaccine_location_data.tail()
To check we can get the list of unique vaccines and crosscheck it with what we had earlier -
vaccine_location_data.vaccines.unique()
We will also have to deal with some missing values in iso_code so that we can merge it with the geodataframe.
Let's get the map data / shapefiles -
import geopandas as gpd
uri_50m = "https://www.naturalearthdata.com/http//www.naturalearthdata.com/download/50m/cultural/ne_50m_admin_0_countries.zip"
uri_110m = "https://www.naturalearthdata.com/http//www.naturalearthdata.com/download/110m/cultural/ne_110m_admin_0_countries.zip"
countries_raw = gpd.read_file(uri_50m)
countries_map = countries_raw.copy()
countries_map.head()
We have a lot of columns here that we do not need. We will drop all that we do not need.
countries_map.columns
#countries_map = countries_map.drop(columns=list(countries_map.columns[:4]) + list(countries_map.columns[5:-1]))
countries_map = countries_map.drop(columns=list(countries_map.columns[:17]) + list(countries_map.columns[18:46]) + list(countries_map.columns[47:-1]))
#countries_map = countries_map.rename(columns={'SOV_A3': 'iso_code'})
countries_map = countries_map.rename(columns={'ISO_A3': 'iso_code'})
countries_map.head()
Let's find out the mismatches of iso_code for the countries that are there in our vaccine dataset and our geodataframe -
vaccine_location_data[~vaccine_location_data.iso_code.isin(countries_map.iso_code)]#['location'].unique()
Let's fix them now -
countries_map.loc[countries_map.NAME == 'France', 'iso_code'] = "FRA"
countries_map.loc[countries_map.NAME == 'Norway', 'iso_code'] = "NOR"
vaccine_location_data.loc[vaccine_location_data.location.isin(['England', 'Wales', 'Scotland', 'Northern Ireland']), 'iso_code'] = "GBR"
FInally let's check once again for any mismatches -
vaccine_location_data[~vaccine_location_data.iso_code.isin(countries_map.iso_code)]#['location'].unique()
For our purposes we can ignore these datapoints. Now let's merge the dataframes and plot the results.
plot_data = countries_map.merge(vaccine_location_data, how='inner', on='iso_code')
plot_data.head()
alt.Chart(plot_data).mark_geoshape().encode(
color='vaccines:N'
).project('equalEarth')
This is a little misleading because countries have multiple vaccines approved for their usages but we will get only one color here. So we will facet our chart based on the vaccine
alt.Chart(plot_data).mark_geoshape().encode(
color='vaccines:N',
#tooltip = ['location'],
facet=alt.Facet('vaccines:N', columns=3)
).properties(width=100, height=100)
Well this is surprising. It should have worked isn't it?
Unfortunately there is a bug with faceting geoshape plots in Vega-Lite that you can follow in this issue. So for that we will try to achieve faceting by concatenating our charts drawn with filtered data.
alt.concat(*(
alt.Chart(plot_data[plot_data['vaccines']==vaccine], title=vaccine).mark_geoshape().encode(
color=alt.value('green'),
#tooltip=['location'],
)
for vaccine in list(plot_data.vaccines.unique())
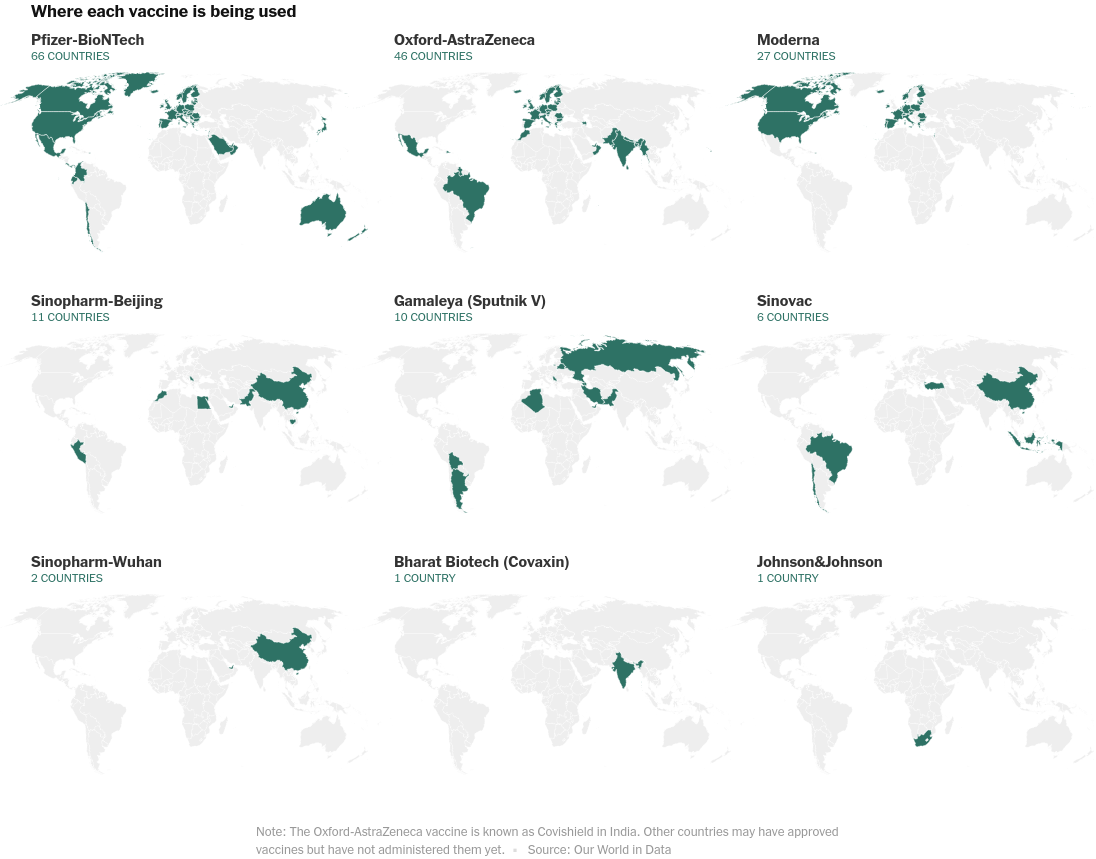
), columns=3, title="Where each vaccine is being used"
)
This is great. Now we will give it some finishing touches -
base = alt.Chart(countries_map[countries_map.iso_code!='ATA']).mark_geoshape(fill='#eee', stroke="#fff", strokeWidth=0.5).project('equalEarth')
alt.concat(*(
base + alt.Chart(plot_data[plot_data['vaccines']==vaccine], title=vaccine, height=200, width=350).mark_geoshape(stroke="#fff", strokeWidth=0.5).encode(
color=alt.value('#2e7265'),
#tooltip=['NAME'],
)
for vaccine in list(plot_data.vaccines.unique())
), columns=3, title="Where each vaccine is being used", spacing=0
).configure_view(strokeWidth=0)